Streaming Total Dynamic Mode Decomposition (i.e. STDMD) is a variant of dynamic mode decomposition.
More...
Public Member Functions | |
| TypeName ("STDMD") | |
| STDMD (const fvMesh &mesh, const word &name, const dictionary &dict) | |
| STDMD (const STDMD &)=delete | |
| void | operator= (const STDMD &)=delete |
| virtual | ~STDMD ()=default |
| virtual bool | initialise (const RMatrix &z) |
| virtual bool | update (const RMatrix &z) |
| virtual bool | fit () |
| virtual bool | read (const dictionary &dict) |
 Public Member Functions inherited from DMDModel Public Member Functions inherited from DMDModel | |
| TypeName ("DMDModel") | |
| declareRunTimeSelectionTable (autoPtr, DMDModel, dictionary,(const fvMesh &mesh, const word &name, const dictionary &dict),(mesh, name, dict)) | |
| DMDModel (const fvMesh &mesh, const word &name, const dictionary &dict) | |
| DMDModel (const DMDModel &)=delete | |
| void | operator= (const DMDModel &)=delete |
| virtual | ~DMDModel ()=default |
| virtual void | reconstruct (const wordList modes) |
| label | nComponents (const word &fieldName) const |
| template<class Type > | |
| bool | nComponents (const word &fieldName, label &nComps) const |
 Public Member Functions inherited from writeFile Public Member Functions inherited from writeFile | |
| writeFile (const objectRegistry &obr, const fileName &prefix, const word &name="undefined", const bool writeToFile=true) | |
| writeFile (const objectRegistry &obr, const fileName &prefix, const word &name, const dictionary &dict, const bool writeToFile=true) | |
| writeFile (const writeFile &wf) | |
| virtual | ~writeFile ()=default |
| virtual OFstream & | file () |
| virtual bool | writeToFile () const |
| virtual bool | canWriteHeader () const |
| virtual label | charWidth () const |
| virtual void | writeCommented (Ostream &os, const string &str) const |
| virtual void | writeTabbed (Ostream &os, const string &str) const |
| virtual void | writeHeader (Ostream &os, const string &str) const |
| virtual void | writeCurrentTime (Ostream &os) const |
| virtual void | writeBreak (Ostream &os) const |
| template<class Type > | |
| void | writeHeaderValue (Ostream &os, const string &property, const Type &value) const |
Additional Inherited Members | |
 Static Public Member Functions inherited from DMDModel Static Public Member Functions inherited from DMDModel | |
| static autoPtr< DMDModel > | New (const fvMesh &mesh, const word &name, const dictionary &dict) |
 Static Public Attributes inherited from writeFile Static Public Attributes inherited from writeFile | |
| static label | addChars = 8 |
 Protected Member Functions inherited from writeFile Protected Member Functions inherited from writeFile | |
| void | initStream (Ostream &os) const |
| fileName | baseFileDir () const |
| fileName | baseTimeDir () const |
| virtual autoPtr< OFstream > | createFile (const word &name, scalar timeValue) const |
| virtual autoPtr< OFstream > | createFile (const word &name) const |
| virtual void | resetFile (const word &name) |
| Omanip< int > | valueWidth (const label offset=0) const |
| void | operator= (const writeFile &)=delete |
 Protected Attributes inherited from DMDModel Protected Attributes inherited from DMDModel | |
| const fvMesh & | mesh_ |
| const word | name_ |
 Protected Attributes inherited from writeFile Protected Attributes inherited from writeFile | |
| const objectRegistry & | fileObr_ |
| const fileName | prefix_ |
| word | fileName_ |
| autoPtr< OFstream > | filePtr_ |
| label | writePrecision_ |
| bool | writeToFile_ |
| bool | updateHeader_ |
| bool | writtenHeader_ |
| bool | useUserTime_ |
| scalar | startTime_ |
Streaming Total Dynamic Mode Decomposition (i.e. STDMD) is a variant of dynamic mode decomposition.
Among other Dynamic Mode Decomposition (DMD) variants, STDMD is presumed to provide the general DMD method capabilities alongside economised and feasible memory and CPU usage.
The code implementation corresponds to Figs. 15-16 of the first citation below, more broadly to Section 2.4.
References:
DMD and mode-sorting algorithms (tags:K, HRDC, KZ, HWR):
Kiewat, M. (2019).
Streaming modal decomposition approaches for vehicle aerodynamics.
PhD thesis. Munich: Technical University of Munich.
URL:mediatum.ub.tum.de/doc/1482652/1482652.pdf
Hemati, M. S., Rowley, C. W.,
Deem, E. A., & Cattafesta, L. N. (2017).
De-biasing the dynamic mode decomposition
for applied Koopman spectral analysis of noisy datasets.
Theoretical and Computational Fluid Dynamics, 31(4), 349-368.
DOI:10.1007/s00162-017-0432-2
Kou, J., & Zhang, W. (2017).
An improved criterion to select
dominant modes from dynamic mode decomposition.
European Journal of Mechanics-B/Fluids, 62, 109-129.
DOI:10.1016/j.euromechflu.2016.11.015
Hemati, M. S., Williams, M. O., & Rowley, C. W. (2014).
Dynamic mode decomposition for large and streaming datasets.
Physics of Fluids, 26(11), 111701.
DOI:10.1063/1.4901016
Parallel classical Gram-Schmidt process (tag:Ka):
Katagiri, T. (2003).
Performance evaluation of parallel
Gram-Schmidt re-orthogonalization methods.
In: Palma J. M. L. M., Sousa A. A., Dongarra J., Hernández V. (eds)
High Performance Computing for Computational Science — VECPAR 2002.
Lecture Notes in Computer Science, vol 2565, p. 302-314.
Berlin, Heidelberg: Springer.
DOI:10.1007/3-540-36569-9_19
Parallel direct tall-skinny QR decomposition (tags:BGD, DGHL):
Benson, A. R., Gleich, D. F., & Demmel, J. (2013).
Direct QR factorizations for
tall-and-skinny matrices in MapReduce architectures.
2013 IEEE International Conference on Big Data.
DOI:10.1109/bigdata.2013.6691583
Demmel, J., Grigori, L., Hoemmen, M., & Langou, J. (2012).
Communication-optimal parallel
and sequential QR and LU factorizations.
SIAM Journal on Scientific Computing, 34(1), A206-A239.
DOI:10.1137/080731992
system/controlDict.functions: DMD1
{
// Mandatory/Optional entries
...
// Mandatory entries (unmodifiable)
DMDModel STDMD;
// Conditional mandatory entries (runtime modifiable)
// Option-1
interval 5.5;
// Option-2
executeInterval 10;
// Optional entries (runtime modifiable)
modeSorter kiewat;
nGramSchmidt 5;
maxRank 50;
nModes 50;
fMin 0;
fMax 1000000000;
nAgglomerationProcs 20;
// Optional entries (runtime modifiable, yet not recommended)
minBasis 0.00000001;
minEVal 0.00000001;
sortLimiter 500.0;
// Mandatory/Optional (inherited) entries
...
}
where the entries mean:
| Property | Description | Type | Reqd | Deflt |
|---|---|---|---|---|
DMDModel | Type: STDMD | word | yes | - |
interval | STDMD time-step size [s] | scalar | cndtnl | executeInterval*(current time-step of the simulation) |
modeSorter | Mode-sorting algorithm model | word | no | firstSnapshot |
nModes | Number of output modes in input frequency range | label | no | GREAT |
maxRank | Max columns in accumulated matrices | label | no | GREAT |
nGramSchmidt | Number of Gram-Schmidt iterations | label | no | 5 |
fMin | Min (non-negative) output frequency | label | no | 0 |
fMax | Max output frequency | label | no | GREAT |
nAgglomerationProcs | Number of processors at each agglomeration unit during the computation of reduced Koopman operator | label | no | 20 |
minBasis | Orthogonal basis expansion threshold | scalar | no | 1e-8 |
minEVal | Min eigenvalue for below eigenvalues are omitted | scalar | no | 1e-8 |
sortLimiter | Max allowable magnitude for mag(eigenvalue)*(number of DMD steps) to be used in modeSorter={kiewat,kouZhang} to avoid overflow errors | scalar | no | 500.0 |
Options for the modeSorter entry:
kiewat | Modified weighted-amplitude scaling method
kouZhang | Weighted-amplitude scaling method
firstSnapshot | First-snapshot amplitude magnitude methodinterval or executeInterval must be available (highest to lowest precedence). While interval allows users to directly specify the STDMD time-step size in seconds, in absence of interval, for convenience, executeInterval allows users to compute the STDMD time-step internally by multiplying itself with the current time-step size of the simulation.modeSorter algorithms of kiewat and kouZhang, a function of  exists where
exists where x is the magnitude of an eigenvalue, and y is the number of STDMD snapshots. This function poses a risk of overflow errors since, for example, if x ends up above 1.5 with 500 snapshots or more, this function automatically throws the floating point error meaning overflow. Therefore, the domain-range of this function is heuristically constrained by the optional entry sortLimiter which skips the evaluation of this function for a given mag(eigenvalue)*(no. of STDMD steps), i.e. x*y, whose magnitude is larger than sortLimiter.| STDMD | ( | const fvMesh & | mesh, |
| const word & | name, | ||
| const dictionary & | dict | ||
| ) |
|
virtualdefault |
| TypeName | ( | "STDMD" | ) |
|
delete |
Implements DMDModel.
Definition at line 865 of file STDMD.C.
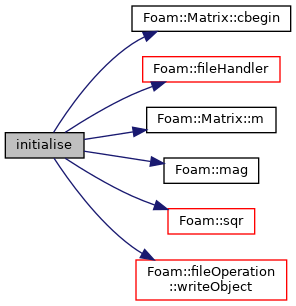
References Matrix::cbegin(), Foam::fileHandler(), Matrix::m(), Foam::mag(), IOobject::NO_READ, IOobject::NO_WRITE, Foam::sqr(), and fileOperation::writeObject().
Implements DMDModel.
Definition at line 921 of file STDMD.C.
References Foam::stringOps::expand(), and Foam::reduce().
|
virtual |
Implements DMDModel.
Definition at line 958 of file STDMD.C.
References UPstream::master(), and Foam::max().
|
virtual |
Implements DMDModel.
Definition at line 778 of file STDMD.C.
References dict, Foam::constant::mathematical::e(), Foam::endl(), MinMax::ge(), Foam::Info, Foam::nl, and Foam::tab.
Copyright © 2011-2018 OpenFOAM | OPENFOAM® is a registered trademark of OpenCFD Ltd.